from sklearn.datasets import load_digits
import seaborn as sns
from sklearn.decomposition import PCA
import pandas as pd
import numpy as np
digits = pd.DataFrame(load_digits()['data'])
classes = load_digits(return_X_y=True)[1]PCA
The principal components are the eigenvectors+eigenvalues of the Covariance matrix of our data.
This is because we are looking for the “Direction of stretching and how much streching happens” regarding the variance of our data.
low_dim_digits = PCA(n_components=2).fit_transform(digits)
sns.scatterplot(x=low_dim_digits[:,0], y=low_dim_digits[:,1], hue=classes)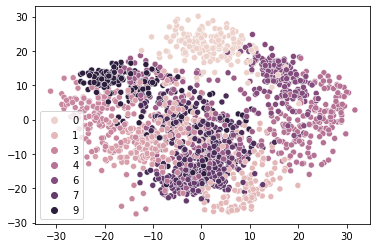
digits_normed = digits - digits.mean()
# compute the covariance matrix
cov_matrix = digits_normed.T @ digits_normed / len(digits_normed) # same as digits_normed.cov()
eigen_values, eigen_vectors = np.linalg.eig(cov_matrix)
eigen_values, eigen_vectors
# Sort eigen values end eigen vectors
sorted_index = np.argsort(eigen_values)[::-1]
sorted_eigenvalue = eigen_values[sorted_index]
sorted_eigenvectors = eigen_vectors[:,sorted_index]
# Select the 2 best
eigenvector_subset = sorted_eigenvectors[:, 0:2]
X_reduced = np.dot(eigenvector_subset.transpose(), digits_normed.transpose()).transpose()
sns.scatterplot(x=X_reduced[:,0], y=X_reduced[:,1], hue=classes)